Project B6: Visualization and analysis of phagophore initiation
Florian Wilfling
Autophagosomes are double membrane organelles which are synthesized de novo to engulf bulky cytosolic material for destruction and can span from 250 nm up to microns in size. Proper autophagosome formation is crucial for cellular homeostasis and malfunction is associated with various human diseases such as neurodegeneration and cancer. A key unanswered question is how an organelle such as an autophagosome can be built from scratch. For autophagosomes different models exist, the most cited model suggests that few vesicles fuse to form a disc or cup-shaped phagophore at the site of cargo recognition, which subsequently expands by direct lipid transfer to engulf the respective cargo1,2. However, due to a lack of resolution direct experimental evidence clarifying the role of different membrane sources in phagophore formation is missing.
In this project, we propose to close this knowledge gap by visualizing the initial stages of phagophore formation in S. cerevisiae by using in situ correlative cryo-electron tomography (cryo-ET).
Our recent study on the characterization of autophagy by cryo-ET demonstrated the feasibility of this approach but was mostly focused on later stages of phagophore formation3. In this project, we plan to specifically target the earliest stages of phagophore initiation using correlative FIB-milling by following a specific strategy: First, we will fluorescently tag the early autophagy marker Atg9, the only transmembrane protein in the autophagy machinery. At the same time, we will localize sites of cargo accumulation, by targeting the autophagy Atg19 and its cargo Ape1 (Figure1). Elusive intermediates of phagophore biogenesis will be enriched by stalling the downstream process using a rapid protein depletion system (auxin-inducible degrons). The high-resolution 3D data obtained in this setup will allow us to analyze multiple aspects of phagophore biogenesis. Specifically, we aim to reveal: 1.) the shape and structure of the early phagophore; 2) which membrane sources and organelles contribute to early phagophore formation (to this end, we will analyze the subcellular landscape at sites of cargo accumulation); and 3) the structural arrangement of the autophagy machinery during phagophore initiation. It has been shown that the early autophagy machinery phase separates in the cytosol which is important for phagophore shaping4. We expect to reveal such phase-separated compartments by mapping ribosome free areas.
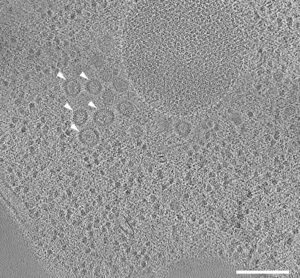
Figure 1: Tomographic slice of stalled autophagosome biogenesis around a selective autophagy cargo. Dotted line indicates an area of Ape1 accumulation. Mutations in the biogenesis pathway led to the accumulation of coated vesicles around the autophagic cargo (arrowheads).
The student working on this project will benefit from ongoing collaborations with labs focusing on the cell biological aspect of autophagosome biogenesis. They will provide different synthetic systems to initiate cargo degradation upon drug addition, which will allow timely controlled initiation of autophagosome formation. The experience from the previous project and exchange with the lead authors of the study will provide the student with expertise and guidance early on. Pipelines for automatic membrane segmentation and data analysis are established. In collaboration, we will use the obtained physical parameters underlying de novo phagophore formation for modelling of the process. Overall, this project will involve a cutting-edge approach to solve a fundamental gap in our understanding how an organelle can be built from scratch and will be well embedded in the iMOL infrastructure.
1. Farre, J. C. & Subramani, S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17, 537-552 (2016). https://doi.org:10.1038/nrm.2016.74
2. Hurley, J. H. & Young, L. N. Mechanisms of Autophagy Initiation. Annu Rev Biochem 86, 225-244 (2017). https://doi.org:10.1146/annurev-biochem-061516-044820
3. Bieber, A. et al. In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc Natl Acad Sci U S A 119, e2209823119 (2022). https://doi.org:10.1073/pnas.2209823119
4. Fujioka, Y. et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301-305 (2020). https://doi.org:10.1038/s41586-020-1977-6
